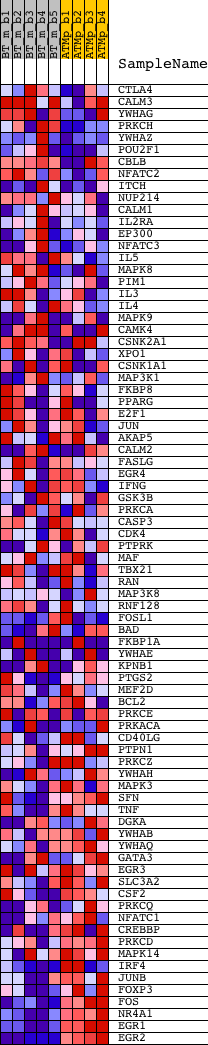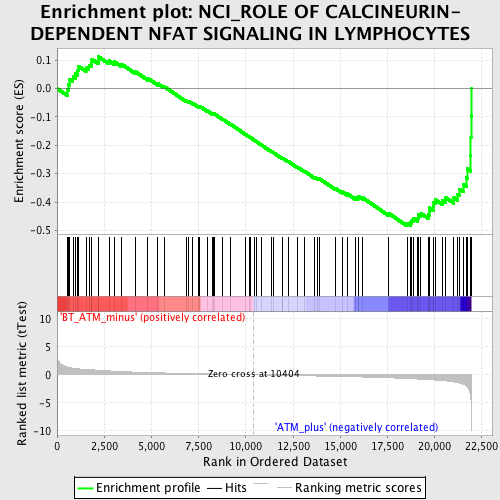
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NCI_ROLE OF CALCINEURIN-DEPENDENT NFAT SIGNALING IN LYMPHOCYTES |
| Enrichment Score (ES) | -0.48443758 |
| Normalized Enrichment Score (NES) | -1.7820044 |
| Nominal p-value | 0.0015772871 |
| FDR q-value | 0.21453731 |
| FWER p-Value | 0.782 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CTLA4 | 1419334_at | 529 | 1.413 | -0.0024 | No | ||
| 2 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 608 | 1.338 | 0.0147 | No | ||
| 3 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 675 | 1.285 | 0.0315 | No | ||
| 4 | PRKCH | 1422079_at 1434248_at | 841 | 1.172 | 0.0421 | No | ||
| 5 | YWHAZ | 1416102_at 1416103_at 1436971_x_at 1436981_a_at 1439005_x_at 1443893_at 1448218_s_at 1448219_a_at | 986 | 1.098 | 0.0525 | No | ||
| 6 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 1100 | 1.059 | 0.0637 | No | ||
| 7 | CBLB | 1437304_at 1455082_at 1458469_at | 1123 | 1.052 | 0.0789 | No | ||
| 8 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 1531 | 0.962 | 0.0751 | No | ||
| 9 | ITCH | 1415769_at 1431316_at 1459332_at | 1688 | 0.928 | 0.0823 | No | ||
| 10 | NUP214 | 1434351_at 1456503_at 1457815_at | 1832 | 0.894 | 0.0896 | No | ||
| 11 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 1843 | 0.892 | 0.1029 | No | ||
| 12 | IL2RA | 1420691_at 1420692_at | 2200 | 0.808 | 0.0991 | No | ||
| 13 | EP300 | 1434765_at | 2203 | 0.808 | 0.1115 | No | ||
| 14 | NFATC3 | 1419976_s_at 1452497_a_at | 2750 | 0.690 | 0.0972 | No | ||
| 15 | IL5 | 1425546_a_at 1442049_at 1450550_at | 3035 | 0.639 | 0.0941 | No | ||
| 16 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 3414 | 0.576 | 0.0857 | No | ||
| 17 | PIM1 | 1423006_at 1435458_at | 4153 | 0.479 | 0.0593 | No | ||
| 18 | IL3 | 1450566_at | 4802 | 0.410 | 0.0360 | No | ||
| 19 | IL4 | 1449864_at | 5329 | 0.360 | 0.0175 | No | ||
| 20 | MAPK9 | 1421876_at 1421877_at 1421878_at | 5667 | 0.330 | 0.0071 | No | ||
| 21 | CAMK4 | 1421941_at 1426167_a_at 1439843_at 1442822_at 1452572_at | 6861 | 0.233 | -0.0439 | No | ||
| 22 | CSNK2A1 | 1419034_at 1419035_s_at 1419036_at 1419037_at 1419038_a_at 1453427_at | 6948 | 0.227 | -0.0443 | No | ||
| 23 | XPO1 | 1418442_at 1418443_at 1448070_at | 7177 | 0.210 | -0.0515 | No | ||
| 24 | CSNK1A1 | 1424827_a_at 1428537_at 1430529_at 1451497_at 1456216_at | 7504 | 0.189 | -0.0635 | No | ||
| 25 | MAP3K1 | 1424850_at | 7535 | 0.186 | -0.0620 | No | ||
| 26 | FKBP8 | 1416113_at | 7954 | 0.158 | -0.0787 | No | ||
| 27 | PPARG | 1420715_a_at | 8208 | 0.139 | -0.0881 | No | ||
| 28 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 8294 | 0.134 | -0.0899 | No | ||
| 29 | JUN | 1417409_at 1448694_at | 8322 | 0.132 | -0.0891 | No | ||
| 30 | AKAP5 | 1447301_at | 8776 | 0.103 | -0.1082 | No | ||
| 31 | CALM2 | 1422414_a_at 1423807_a_at | 9204 | 0.077 | -0.1266 | No | ||
| 32 | FASLG | 1418803_a_at 1449235_at | 9986 | 0.027 | -0.1619 | No | ||
| 33 | EGR4 | 1449977_at | 10167 | 0.015 | -0.1699 | No | ||
| 34 | IFNG | 1425947_at | 10234 | 0.011 | -0.1728 | No | ||
| 35 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 10461 | -0.003 | -0.1831 | No | ||
| 36 | PRKCA | 1427562_a_at 1443069_at 1445028_at 1445395_at 1446598_at 1450945_at | 10542 | -0.008 | -0.1866 | No | ||
| 37 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 10836 | -0.027 | -0.1996 | No | ||
| 38 | CDK4 | 1422441_x_at | 11336 | -0.058 | -0.2215 | No | ||
| 39 | PTPRK | 1423277_at 1423278_at 1431680_a_at 1441276_at 1441277_s_at 1441644_at 1443491_at 1443699_at 1444763_at | 11448 | -0.065 | -0.2256 | No | ||
| 40 | MAF | 1435828_at 1437473_at 1444073_at 1447849_s_at 1447945_at 1456060_at | 11924 | -0.094 | -0.2459 | No | ||
| 41 | TBX21 | 1449361_at | 11939 | -0.095 | -0.2450 | No | ||
| 42 | RAN | 1443131_at 1446819_at 1447478_at 1460551_at | 12231 | -0.112 | -0.2566 | No | ||
| 43 | MAP3K8 | 1419208_at | 12729 | -0.144 | -0.2771 | No | ||
| 44 | RNF128 | 1418318_at 1449036_at | 13088 | -0.166 | -0.2910 | No | ||
| 45 | FOSL1 | 1417487_at 1417488_at | 13631 | -0.203 | -0.3126 | No | ||
| 46 | BAD | 1416582_a_at 1416583_at | 13788 | -0.213 | -0.3165 | No | ||
| 47 | FKBP1A | 1416036_at 1438795_x_at 1438958_x_at 1448184_at 1456196_x_at | 13886 | -0.221 | -0.3175 | No | ||
| 48 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 14726 | -0.281 | -0.3515 | No | ||
| 49 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 15097 | -0.307 | -0.3637 | No | ||
| 50 | PTGS2 | 1417262_at 1417263_at | 15377 | -0.327 | -0.3714 | No | ||
| 51 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 15790 | -0.359 | -0.3848 | No | ||
| 52 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 15936 | -0.372 | -0.3856 | No | ||
| 53 | PRKCE | 1437860_at 1437861_s_at 1442609_at 1444649_at 1449956_at 1452878_at | 15965 | -0.375 | -0.3811 | No | ||
| 54 | PRKACA | 1447720_x_at 1450519_a_at | 16164 | -0.390 | -0.3842 | No | ||
| 55 | CD40LG | 1422283_at | 17543 | -0.526 | -0.4391 | No | ||
| 56 | PTPN1 | 1417068_a_at 1438670_at | 18535 | -0.658 | -0.4743 | Yes | ||
| 57 | PRKCZ | 1418085_at 1449692_at 1454902_at | 18718 | -0.686 | -0.4720 | Yes | ||
| 58 | YWHAH | 1416004_at | 18791 | -0.697 | -0.4645 | Yes | ||
| 59 | MAPK3 | 1427060_at | 18852 | -0.706 | -0.4564 | Yes | ||
| 60 | SFN | 1448612_at | 19108 | -0.749 | -0.4565 | Yes | ||
| 61 | TNF | 1419607_at | 19120 | -0.750 | -0.4454 | Yes | ||
| 62 | DGKA | 1418578_at | 19270 | -0.779 | -0.4402 | Yes | ||
| 63 | YWHAB | 1420878_a_at 1420879_a_at 1420880_a_at 1436783_x_at 1438708_x_at 1455815_a_at | 19654 | -0.852 | -0.4445 | Yes | ||
| 64 | YWHAQ | 1420828_s_at 1420829_a_at 1420830_x_at 1432842_s_at 1437608_x_at 1454378_at 1460590_s_at 1460621_x_at | 19700 | -0.861 | -0.4333 | Yes | ||
| 65 | GATA3 | 1448886_at | 19704 | -0.862 | -0.4201 | Yes | ||
| 66 | EGR3 | 1421486_at 1436329_at | 19914 | -0.912 | -0.4156 | Yes | ||
| 67 | SLC3A2 | 1425364_a_at | 19929 | -0.915 | -0.4021 | Yes | ||
| 68 | CSF2 | 1427429_at | 20032 | -0.940 | -0.3922 | Yes | ||
| 69 | PRKCQ | 1426044_a_at | 20416 | -1.029 | -0.3939 | Yes | ||
| 70 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 20589 | -1.073 | -0.3852 | Yes | ||
| 71 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21006 | -1.257 | -0.3848 | Yes | ||
| 72 | PRKCD | 1422847_a_at 1442256_at | 21204 | -1.377 | -0.3725 | Yes | ||
| 73 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 21320 | -1.472 | -0.3551 | Yes | ||
| 74 | IRF4 | 1421173_at 1421174_at | 21520 | -1.685 | -0.3381 | Yes | ||
| 75 | JUNB | 1415899_at | 21672 | -1.992 | -0.3143 | Yes | ||
| 76 | FOXP3 | 1420765_a_at 1457082_at | 21728 | -2.176 | -0.2832 | Yes | ||
| 77 | FOS | 1423100_at | 21901 | -3.644 | -0.2348 | Yes | ||
| 78 | NR4A1 | 1416505_at | 21917 | -3.996 | -0.1737 | Yes | ||
| 79 | EGR1 | 1417065_at | 21922 | -4.993 | -0.0968 | Yes | ||
| 80 | EGR2 | 1427682_a_at 1427683_at | 21930 | -6.298 | 0.0002 | Yes |